Erika Update #10: Honestly, it’s often faster to do it by hand
A cautionary tale of how scripting is not always worth it
When scientists from other fields enter biology they are often surprised by the remarkable amount of repetitive work. I had this reaction too. As a former computer science major, I fancied myself a person who knew how to accelerate boring/repetitive tasks, and gleefully took to writing scripts to ‘accelerate’ many aspects of the computer work. Spoiler: it wasn’t worth it.
Case in point, I happened to need to test out 60 different specific point mutations of a protein I was working with (see Erika Update #10, linked below). To do so, I faced the prospect of clicking and typing 60 times to make 60 different plasmid maps on Benchling, clicking a whole bunch to design each of 60 sets of primers, then using my hands to clone 60 plasmids (a lot of pipetting), doing all the logistics to sequence 60 plasmids and store the glycerol stocks in the fridge, and then use my fingers to do 60 plaque assays and then take 60 images with a camera and then use my fingers to manually make a figure of all the results! The indignity!
I promptly set to work scripting a lot of these bits. I got the Benchling API working and wrote a script that will make a plasmid map and design primers for me based on a desired point mutation string like “A207G”. The pipetting I did by hand because Ahmed doesn’t believe in ordering oligos in plates (“they’re not reusable”), meaning that I had to manually resuspend 120 primers, set up 60 PCRs, etc, all partially with a single-channel. Once the cloning was done, I got a bit stuck on doing sequence analysis. I was on a mission to eliminate all clicking! I really wanted Genewiz to send the sequencing results to an email that would automatically align them to the proper benchling plasmid and just text me to say which plasmid had exactly validated. Unfortunately there was no way to upload sequencing results through the Benchling API, so I ended up with this weird hybrid system that stored plasmid maps on Benchling, but aligned and saved sequencing results locally. Then we come to the data analysis step. I ended up with 60 pictures of plaque assays (quarter agar plates with little polka dots on them). I wanted to compile a little swatch of each of the plates, and I just couldn’t bear the thought of doing it by hand, so I wrote this whole image analysis script that automatically identifies the circular plate in the image and cuts out a swatch of it.
Was it worth it? No it sure wasn’t! It probably took a week to write all this, and I definitely could have done it faster by hand. On top of that, I reused very little of the code I had written. Everything I wrote that was connected to Benchling was absolutely not worth it. The main issue is that the Benchling API just wasn’t complete enough to allow you to automate all actions you might take as a human. I was in the habit of storing my plasmid sequencing information in the alignments of the corresponding Benching map, and if an automated version of cloning forced me to change my workflow, it was out! In addition, most of the cloning I needed to do wasn’t point mutations, it was more complex and bespoke stitching together of two or three pieces, and so it didn’t fit into this nice little string → plasmid interface I had written.
Honestly the most damning part of this was the image analysis. It didn’t work all that well. You can see in the update that it does a pretty-good but not-perfect job of cutting swatches out of the middle of the quarter of each plate. Is it possible to do the image analysis better? For sure! I just didn’t have time to futz with it.
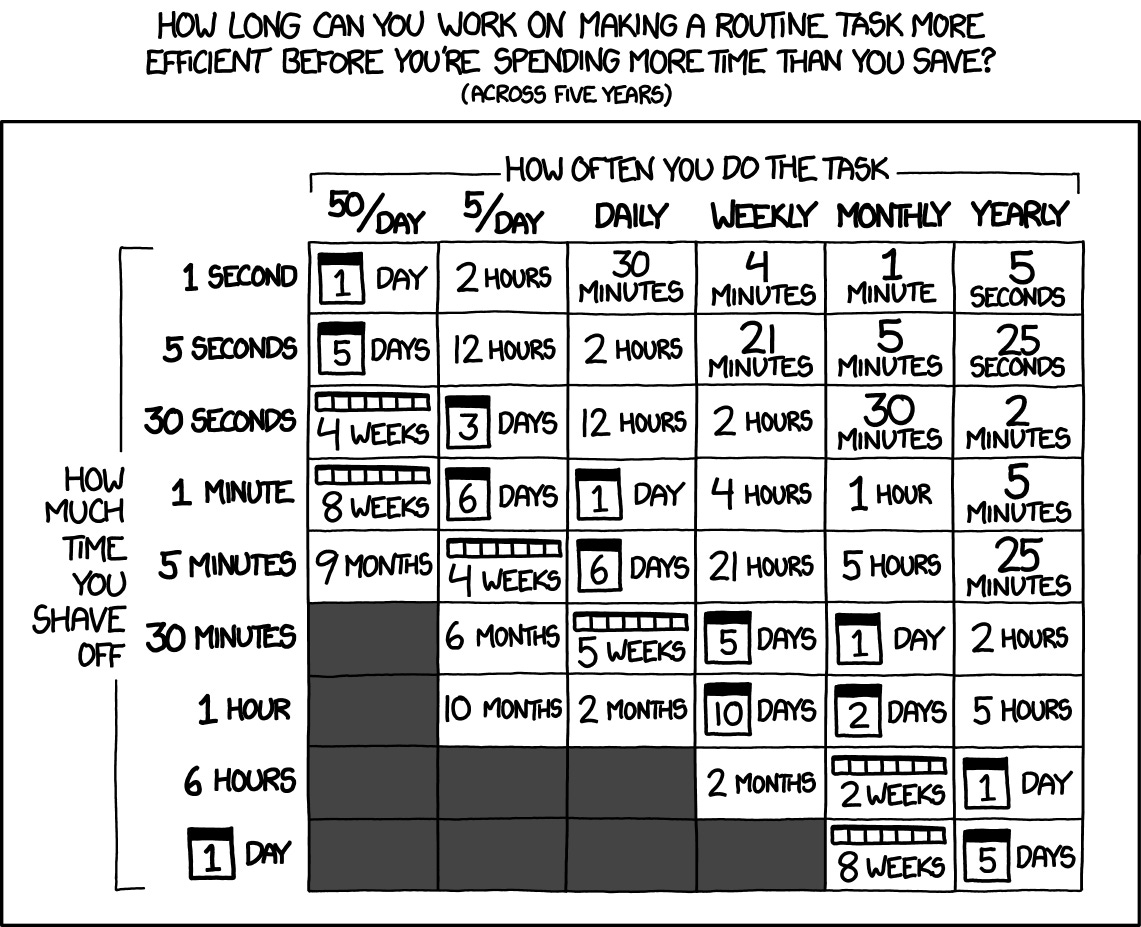
The exception was that I did expose the USER primer design tool as its own endpoint, expanded on it over the years, and all together me and other lab members got a lot of use out of it. That was net positive. This tool ended up being worth it because I actually adopted it, and so it saved a little bit of time over a long time for many people, and thus passes the XKCD test:
Erika Update #10
Here’s Erika Update #10 2018 8 1 - pIII residue specificity
In this update, I’m searching for locations in the protein that can’t be mutated. I’m hoping to find these because then I can use them as sensors that detect not just whether or not a specific qtRNA works at all, but also whether or not it incorporates a specific amino acid.
In order to do this, Ahmed and I opened up a crystal structure of the pIII virus tail fiber protein and eyeballed it to pick which residues to test. This was the first time I’d seen someone open a crystal structure and ‘know’ things about residues - Ahmed has some real chemical intuition about amino acids, which was very cool! Much later, when I did my postdoc in the Baker lab, I encountered this whole world of people who are a level up with their understanding of proteins, and can tell you things not only about the individual residues, but about how the whole protein holds together.
Where’d it end up?
Nowhere! Although this data does get cited in a much later Erika-update.
And to orient you
I think pretty much everyone can get something out of reading these blog posts + updates. Here’s some additional notes customized for you, depending on your interests + background:
You’re a non-scientist: This update is a cautionary tale of how overcomplicating things is a bad idea - done is better than fancy!
You’re a student doing a PhD/undergrad research/etc: I know a lot of people who have written their own cloning code stack. I know very few people who think it was worth it. 😜
You have ideas for reforming publishing: This is some nice data! Someone else who does PACE will surely need this data one day. This is a nice example of something that might make for a good micropublication.
You’re a fellow PACE nerd: Ahmed is a big believer in looking at plaque morphology. It’s true that plaque morphology can sometimes tell you interesting things, but in this case I actually think it would have been better to just do enrichment assays on all of them - it’s more quantitative. Also, there’s a really lovely new cryo-EM structure of pIII now, check it out!
Want more?
If you want to follow along with this project, you can get updates by signing up through substack, or following me on linkedin or twitter.
If you have ideas for what I should cover in the blog post, suggestions for vocabulary to define, questions about the science, or other comments, please do reach out by twitter DM - I’d love to hear from you!





Have you ever tried using something like emerald cloud lab? I would think that's the kind of API we need in order to actually automate all of these protocols, then all that is required is a hub for sharing protocols on such a platform
>I really wanted Genewiz to send the sequencing results to an email that would automatically align them to the proper benchling plasmid and just text me to say which plasmid had exactly validated.
That would be awesome and save me a lot of time. Hopefully Benchling can improve their API.