Biology is a DNA-programmable. In computer science you might use a compiler to convert instructions into machine code your computer can execute; in biology you convert instructions into a custom piece of DNA that the cell can execute. As the basis of all biological engineering, it’s really valuable to keep good track of all of the “DNA programs” you’ve written. Here’s how I wish it worked:
Every published paper would present clear nomenclature for naming the custom pieces of DNA they use that would make it clear which DNA was used to generate each result.
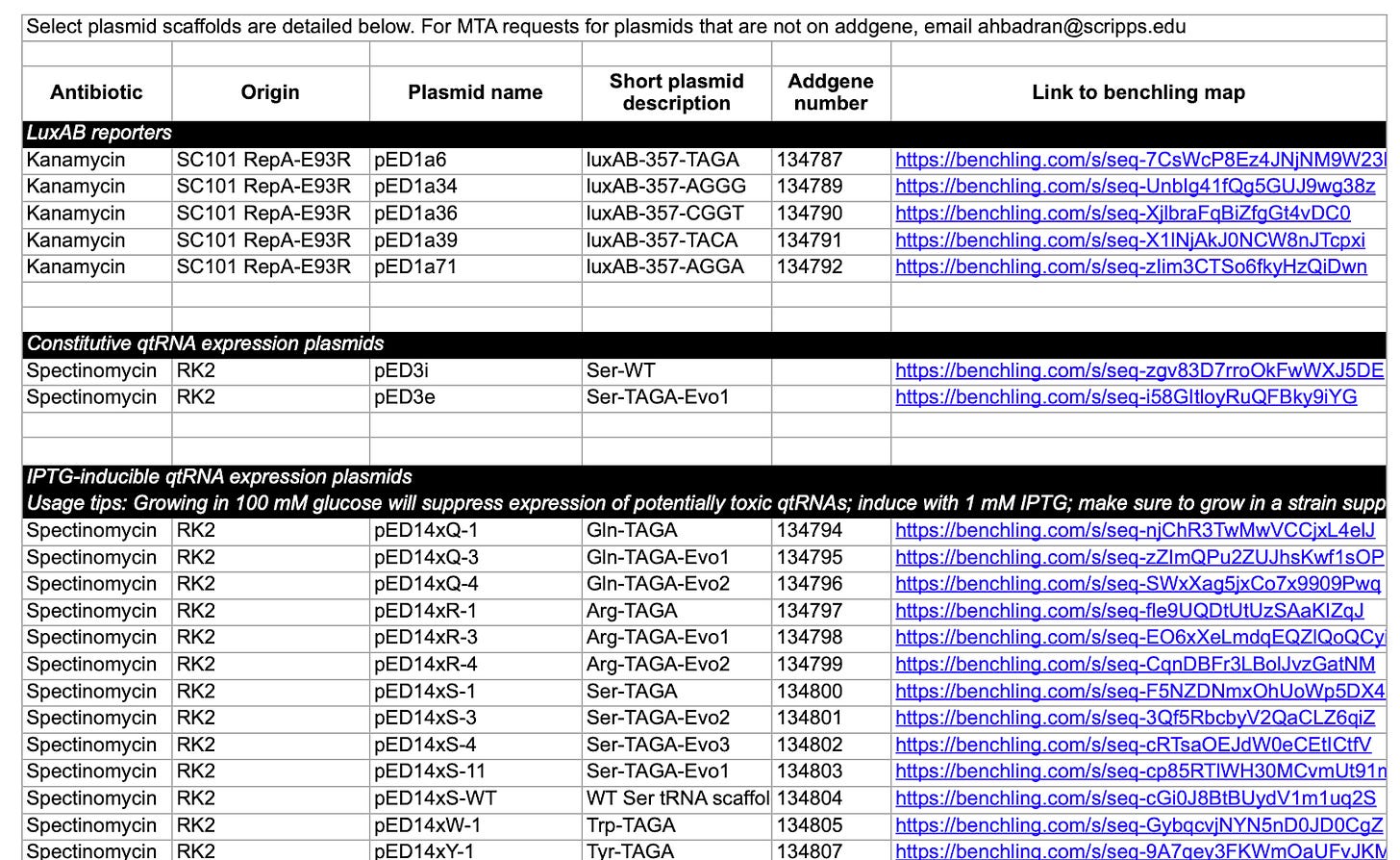
The supplemental information would include a link to every custom DNA that was used in the study. You would just click a link and it would show you the sequence of the DNA, information about how it was constructed, and have sequencing results attached.
We would have tools to ‘diff’ two different pieces of DNA and highlight the differences. Ya know, like Github but for DNA.
I’m basically describing how I do things. (1) and (2) are what I do when I write papers. For example, in this paper you can just check out the “Extended Supplement” spreadsheet in “Source Data” to find links to every plasmid map used to generate every datapoint in every figure in the paper. (3) doesn’t exist, as far as I know.
Instead, that is not at all how it works!
Here’s what DNA management looks like today:
When you read papers there isn’t clear nomenclature. Figuring out what DNA was used in each experiment usually involves a careful reading of the methods section, the supplemental information, and sometimes and email to the author. Many papers don’t even attempt to make the exact DNA sequences they use available.
People do sometimes list their DNA constructs… in PDF or JPEG format. I could not make this up! They literally put their DNA sequences in supplemental information in a format that can’t be copy pasted. I have spent many an hour transcribing and double-checking DNA out of the supplemental information of papers.
There are many different file formats for DNA. They all succeed in listing the sequence of the DNA, so big win there! Unfortunately, the annotations are also important, and the annotations often get lost through file conversion. Why did the author put these pieces together? What does the author think the function is of these different bits? Can I delete this? These are all questions that are assisted by annotations. Annotations often get lost or deleted because they’re not handled smoothly across different software and file formats. For example, when you send in DNA to Addgene, it will usually delete most of the annotations.
When you get DNA from someone else, often you need to do some ‘detective work’ to figure out what’s what. I often align a suite of antibiotic resistance markers and origins to figure out what the backbone is of unmarked plasmids. I dig around looking for tRNAs using secondary structure prediction. I use the RBS/promoter calculator to try to re-annotate promoters whose annotations have been long lost, like I did in Erika Update #5, linked below.
Sometimes instead of being not annotated, maps are mis-annotated. My postdoc once came to me and was like, “heyyy, you know how all your plasmids use the SD8 ribosome binding site? It’s not SD8 at all…”
DNA management is a genuinely tough problem
DNA management is a bit of a jumble today because it actually touches up against some fundamental basic science questions. We often don’t know why things work in biology! Part of why it’s not possible to ‘diff’ to different sequences of DNA is because it’s a genuinely tough biological problem to determine whether or not two pieces of DNA have the same function. This has ramifications for DNA map management, amongst other weighty topics like biosecurity.
If you’re beginning with a natural sequence of DNA and making modifications, best practice is usually to modify as little as possible. They prefer the wildtype sequence over many other possible sequences unnecessarily. In an engineering context, this has real practical implications. I’m often hesitant to touch areas of the DNA that are unannotated. Is there something important there? Point mutations can change plasmid copy number. Regulatory elements aren’t always obvious. What does this random bit of space in between these two genes do? If I don’t know I’m not touching it! Chris Voigt has a good phrase for this bias: he says that many people “worship the wildtype” sequence.
As evidenced by Erika Update #5, my paranoia is sometimes in vain. In this instance, I had discovered that a promoter in a piece of DNA I was using was mis-annotated. I went to some trouble to both fix the annotation and propagate that change to how I was making modifications to the DNA down the line. It didn’t matter. Sometimes it does.
Fortunately, things are looking up. Benchling is a big step forward in terms of modern software to visualize and share DNA maps. Nanopore sequencing providers like Plasmidsaurus and Primordium for upping the game with the technical capability to nicely sequence large DNA constructs, and by default when they auto-annotate the map when they send you sequencing results.
Erika Update #5
Here’s Erika Update #5 - 2018 4 7 - inducible tRNA backbone.pdf
In this update, I’m finally getting tools in place to deal with all the pesky toxicity that was making my engineered tRNAs tough to work with. Up to this point I was having a lot of trouble working with these engineered tRNAs because some of them are very toxic, so toxic that you can’t make them reliably or handle them reliably. On the first page of the update, I talk about testing a few different options for an inducible promoter. The idea here is that instead of expressing a gene all the time, you can encode the gene but not express it until adding a small molecule.
On the third page, I dive in to tuning my circuit for PACE. I’m tweaked the promoter strength in my gene circuit so that the difference between robust phage growth (nice dark polka dots) and no phage growth depends on use of both an engineering tRNA. I still can’t get it to also rely on the engineered ribosome though…
Where’d it end up?
None of the data in this update was published, although the IPTG-inducible tRNA expression plasmid ended up getting used throughout the project, and my lab still uses it to this day.
And to orient you
I think pretty much everyone can get something out of reading these blog posts + updates. Here’s some additional notes customized for you, depending on your interests + background:
You’re a non-scientist: I’m trying to make a ‘dimmer knob’ for my bacteria that I can use to turn on and off my engineered tRNA. I tested a couple different ways to make a dimmer knob, and ended up choosing one that’s turned on and off by how much IPTG I feed the bacteria. IPTG is a type of sugar.
You’re a student doing a PhD/undergrad research/etc: A lot of the inducible promoters come from sugar metabolism, where the presence of a certain type of sugar will start expressing genes that are used to digest it. I tested two different types of sugar-inducible promoters, one that’s induced by rhamnose and another by IPTG. I ended up going with the IPTG one although they’re pretty similar.
You have ideas for reforming publishing: The IPTG inducible promoter was actually a new design that Ahmed came up with, and I don’t think anyone else uses it. There’s no way to publish useful parts like this.
You’re a fellow PACE nerd: The proA/B/C/D series is great for circuit tuning. Phage enrichment is a good benchmark, but strong plaques are practically speaking super useful to have. Often the difference between plaquing and not plaquing is as simple as changing promoter strength.
Want more?
If you want to follow along with this project, you can get updates by signing up through substack, or following me on linkedin or twitter.
If you have ideas for what I should cover in the blog post, suggestions for vocabulary to define, questions about the science, or other comments, please do reach out by twitter DM - I’d love to hear from you!




> We would have tools to ‘diff’ two different pieces of DNA and highlight the differences. Ya know, like Github but for DNA.
I remember the [Vanderbilt 2014 iGEM team](https://2014.igem.org/Team:Vanderbilt_Software/Project/Home) (which I think was [basically](https://github.com/cosmicexplorer/darwin) "Danny McCalahan") tried to make a simple git-based version control tool for DNA... In some ways I feel like we've made a lot of progress since then (I see a lot of teams using Benchling, and wonder if we'll see more people experimenting with other LIMS / lab notebook service software).
>Many papers don’t even attempt to make the exact DNA sequences they use available.
Often because they want to keep a (perceived) competitive advantage. I once emailed some authors and asked for expression vector sequences for BMP15 and GDF9, and they refused to share them.
I think journals should require sharing sequences as a condition of publication. It's really not that hard.